As the primary drivers of biological functions, proteins are central to understanding and treating disease, especially in oncology. Here, the challenge involves targeting specific cancer-related proteins that are either overexpressed, underexpressed, or mutated.
Proteins — including antibodies, growth factors, and peptides — also have emerged as a potent modality for cancer treatment[1]. These therapies, actively targeting tumor cells, constitute a significant proportion of currently approved cancer treatments, offering a more precise approach compared to traditional small molecule chemotherapeutics[2].
With advancements in gene expression technology, improved protein engineering, and refined analytical tools, this area of medicine is expected to expand even more.
We present an analysis of a large, multi-dimensional proteomic dataset to unravel the intricate landscape of cancer proteomics. We also examine approximately 100 antibody-based cancer therapies currently in clinical settings, aiming to identify opportunities for innovation in protein-based therapeutic development.
Mapping the diversity of cancer proteins
Understanding and harnessing the structural and functional characteristics of cancer targets requires the high-quality expression and purification of these proteins using recombinant expression technology[3]. This facilitates the exploration of disease pathways, streamlines the identification of promising drug candidates, and validates therapeutics pre-clinically. Access to target proteins ensures each research phase is grounded in accurate, reliable biological data[4].
With the advent of large, multi-dimensional databases, we now have access to an array of data encompassing sequence, functional, and clinical information derived from human cancers.
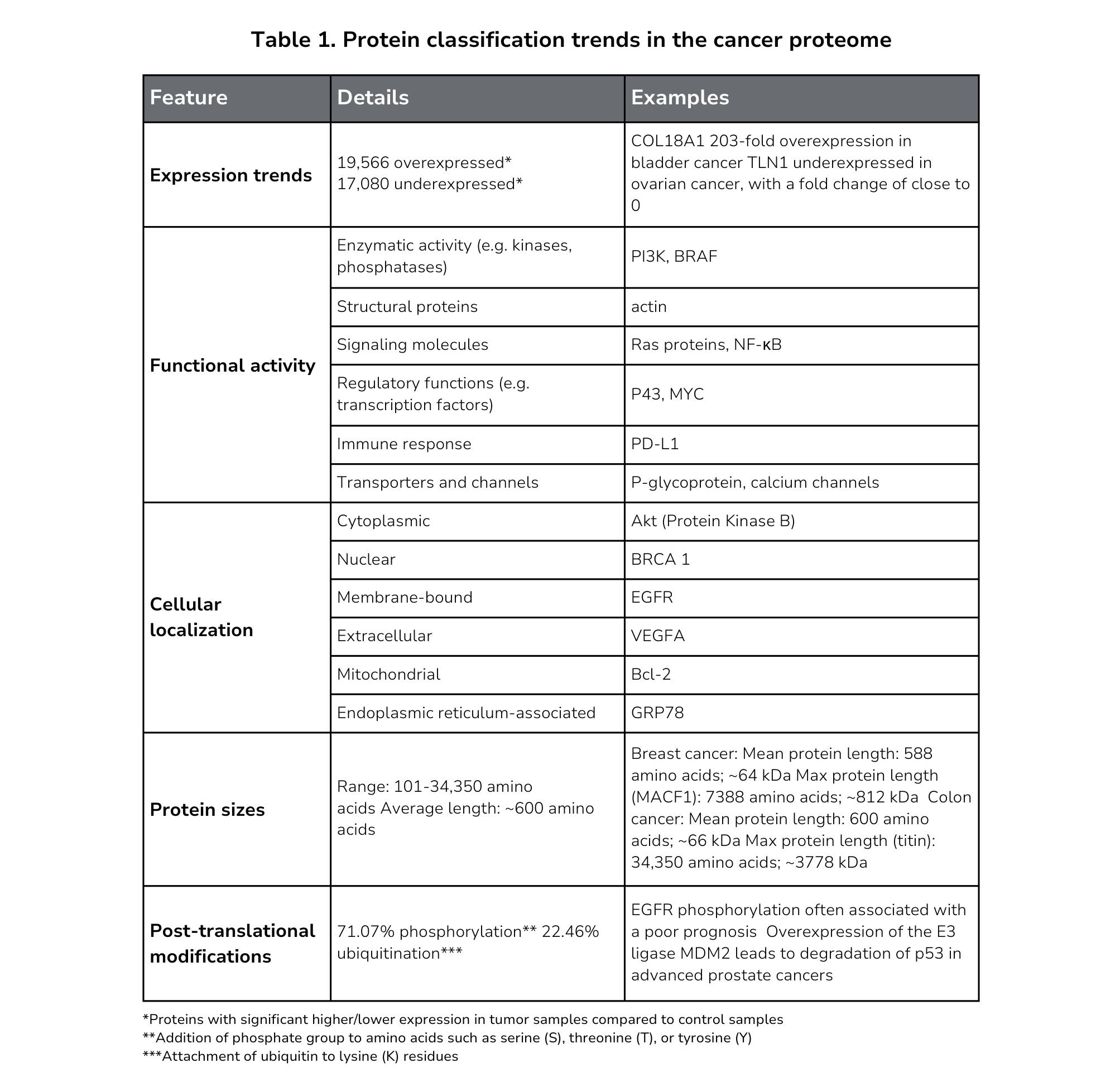
One example is the CancerProteome database featuring over 36,000 proteins from patient samples across 21 cancers[5]. The database reveals a remarkable diversity among potential cancer targets, including a wide range of protein sizes, functions, and cellular locations (Table 1).
Novel strategies for tumor targeting
Despite the expansive and varied cancer proteome research increasingly focuses on specific protein families for next-generation therapeutic strategies. Recent advancements in cancer treatment include:
Targeting the tumor microenvironment
- Angiogenesis inhibitors targeting receptors like NRP1 and NRP2 disrupt the tumor vasculature and reduce drug resistant[6]
- Reprogramming tumor-associated macrophages to inhibit tumor growth[7]
Reactivating immune responses to cancer
- Targeting immune checkpoint proteins such as PD-1, CTLA-4, LAG-3, and TIGIT[8]
Disrupting signal transduction pathways
- Blocking cellular communication networks such as the Wnt/β-catenin, Hedgehog, NOTCH, and cell cycle pathways[9]
Challenges of probing the cancer proteome
Synthesizing cancer proteins that closely resemble their native state in the human body is a pivotal element in propelling drug discovery. However, achieving high-fidelity replication (with functional folding and post-translational modifications (PTMs)) is a challenge, particularly with high-throughput expression methods.
Challenges in recombinant protein expression include:
- Proteins with complex 3D structures, such as kinases, can be challenging to synthesize due to the need for specific folding conditions
- Membrane-associated proteins are difficult to isolate due to their hydrophobic regions and complex topology
- Expressing larger proteins that are prone to misfolding or degradation
- Synthesizing proteins with extensive PTMs can be challenging to replicate in vitro, affecting their functionality
Innovative synthesis strategies are crucial to meet the growing demand for cancer-specific proteins.
Future progress hinges on expression technologies that simplify protein production and enable high-throughput screening of diverse reaction parameters, gene variants, and supplements. These technologies can help maintain cost-efficiency while accelerating drug development[10].
Protein-based cancer therapeutics
Protein-based cancer therapeutics offer several advantages over small molecules for treating cancer. They have a larger binding surface area, are customizable for various targeted therapeutic mechanisms, and, because they remain in the blood circulation longer, require less frequent doses[11].
Emerging peptide-based therapies, by attacking cancer cells while sparing healthy ones, represent a promising branch of these therapies[12]. Peptides such as leuprolide, a somatostatin analog used to treat prostate cancer, can target receptors on tumors, deliver payloads, and inhibit cell signaling[13,14].
Antibody therapeutics are predominant in cancer therapy, with most clinical-stage antibodies approved for oncology applications[13,15].
Therapeutic targets:
- The most commonly targeted molecules are PD-1, PD-L1, HER2, CD20, and EGFR, underscoring these targets' importance in cancer therapeutics and the opportunity for new entrants that can offer differentiated mechanisms or improved efficacy.
Antibody format and optimization strategies:
- A dominant 94.68% of clinical-stage antibodies for cancer are in the IgG format, highlighting the industry's preference for this format due to its well-understood structure and function.
- The majority (86%) of these antibodies are full-length, indicating a strong industry trend towards using complete antibody structures, which may offer advantages in binding affinity and specificity. Formats like scFv, VHH-Fc, Fabs are less prevalent.
- There's a significant focus on monoclonal antibodies (83%), known for their specificity and consistency, while bispecific antibodies (16%) are gaining traction, offering the potential to target multiple antigens simultaneously.
- Approximately half of the antibodies are Fc-engineered, indicating a significant interest in modifying effector functions and pharmacokinetics to enhance therapeutic effectiveness. The S228P modification is the most common among Fc-engineered antibodies.
Manufacturing systems:
- Chinese hamster ovary (CHO) cells are the predominant system for antibody expression, reflecting their reliable use in large-scale production. Alternative systems like NS0 cells, FUT8−/− CHO cells (Potelligent® Technology), and Sp2/0 cells are less commonly used.
Challenges in protein-based therapeutic production
Manufacturing protein-based cancer therapies, especially on a large scale, presents significant challenges due to their complex nature.
Antibody candidates in development often need to undergo extensive engineering and optimization steps before they can be safe, effective, and manufacturable[16].
Antibodies can also encounter solubility and stability issues, which can hinder their production and formulation for therapeutic use[16]. To enhance stability and solubility, proteins are sometimes modified using techniques like PEGylation. However, this adds complexity to the manufacturing process.
The considerable size (around 150 kDa) and intricate structure of antibodies introduce complexities in their manufacturing process, especially when compared to smaller, less complex molecules. This complexity often hampers the ability to consistently achieve high-quality production and maintain optimal yield.
Innovations expediting discovery
Whether in the discovery of novel cancer targets or in the development of therapeutics, rapid and efficient engineering cycles are pivotal. Technologies that facilitate the combinatorial testing of diverse cancer-specific proteins, sourced from metagenomic data, computationally modeled, or bioengineered, greatly enhance the efficiency of this process.
These protein synthesis technologies should be:
- versatile and customizable, to synthesize a wide array of challenging proteins
- scalable, to efficiently generate vast libraries of protein variants
- reliable, synthesizing high-quality proteins with precision and accuracy
Additionally, emerging technologies powered by artificial intelligence now enable the de novo synthesis of computationally designed proteins, directly from sequence lists, effectively bypassing the constraints of traditional protein expression methods[17]. By utilizing high-throughput, cell-free synthetic biology, these platforms overcome the limitations of in-cell testing, enabling the rapid synthesis and evaluation of extensive protein libraries[10].
Ultimately, these innovative approaches offer a deeper and more nuanced exploration of proteins, hastening the development of more effective and personalized cancer treatments.