Picture a future where high-quality therapeutic proteins are manufactured in hours instead of days. Cell-free protein synthesis (CFPS) platforms enable reliable protein synthesis at any scale, driving biotherapeutic discovery and production. This protocol uses cellular lysates to extract the transcription-translation machinery and energy components, redirecting them to produce a target protein.
As CFPS becomes more affordable, various systems have emerged, necessitating careful selection to optimize protein yield and quality[1]. Here, we explore the current CFPS landscape, factors influencing system choice, and the role of strain engineering in the evolution of CFPS.
Prokaryotic CFPS Systems
Prokaryotes couple transcription and translation through ribosomes associating with mRNA before transcription is completed[2]. This simultaneous process minimizes naked mRNA accumulation and resource depletion, enhancing efficiency during protein synthesis[3]. The potential was first realized over 60 years ago with an E. coli-based CFPS system, leading to the synthesis of various therapeutically relevant proteins[4].
Since then, multiple prokaryotic-based CFPS platforms have generated an array of biopharmaceutical proteins, including:
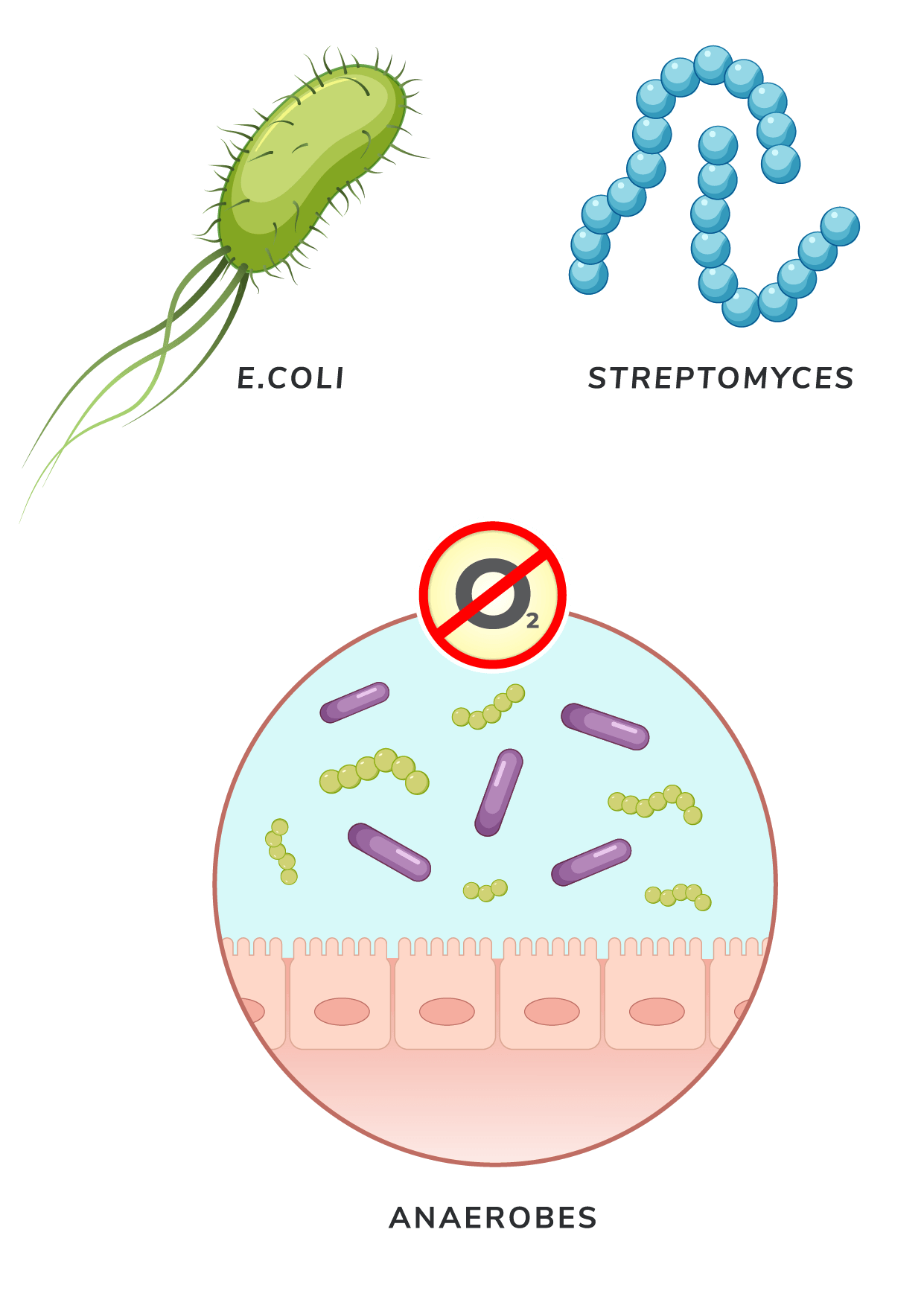
E. coli
- The first bacterial species used for CFPS, E. coli, is cost-effective due to its high growth rates, aiding in the discovery of SARS-CoV-2 antibodies and Shiga toxin characterization[5,6].
Vibrio spp.
- Fast-growing Vibrio natriegens synthesizes proteins efficiently, supported by 3-PGA as an energy source, making it ideal for producing nonribosomal peptides and natural products such as antibiotics[7-9].
Streptomyces
- Streptomyces has been a major source of natural products in medical care, from antibiotics to crop-protecting agents. Cells in this genus harbor enzyme-coding genes that confer strong capabilities to be a CFPS system[10]. Most notably, species in this genus can express phosphopantetheinyl transferases that activate carrier proteins involved in the biosynthesis of complex natural products like antibiotics and polyketides[11].
Anaerobes
- The emerging use of anaerobic bacteria in CFPS is opening new possibilities for sustainable production[12]. Recently, companies like LanzaTech have announced the development of commercial-scale bioreactors that use anaerobes to metabolize gas emissions to produce ethanol and other compounds.
In addition to these species, multiple strains within each confer distinct advantages for CFPS. For example, E. coli DE3 strains eliminate the need to add polymerases by inducing sufficient T7 RNA polymerase expression[13].
Even so, most prokaryotes produce a limited number of post-translational modifications, making them less suitable for producing complex proteins with glycosylation or disulfide bonds, which are essential for their function and therapeutic efficacy[14].
Eukaryotic CFPS Systems
The challenge of producing proteins with post-translational modifications in prokaryotic systems has led to the increased use of eukaryotic CFPS systems. Several eukaryotic systems have been developed and tailored for protein synthesis, including:
Spodoptera frugiperda 21 (Sf21)
- Despite being a pest, the fall armyworm's cell extracts are valuable for producing integral membrane proteins[15,16].
Wheat germ
- Wheat germ is used to produce proteins at the milligram scale. Although raw extracts are challenging to process, refined protocols minimize RNase activity and reduce yield variation. These improvements have enabled wheat germ systems to produce purified recombinant proteins and vaccine candidates, such as those for malaria[17-21].
Tobacco BY-2
- Tobacco Bright Yellow-2 (BY-2) lysates offer high translational activity, quick preparation, and scalability to milligram and gram levels of protein. BY-2 cell lysates were used to produce the first FDA-approved recombinant pharmaceutical protein, Taliglucerase alfa (Elelyso®)[22].
Human cell cultures
- Various human cell lines, such as HEK293, HeLa, and CHO cells, are used to express diverse proteins using an in vitro coupled transcription/translation system. Like Sf21, human-based systems have ER-derived microsomes that facilitate the production of integral membrane proteins[23].

Factors to Consider When Selecting a CFPS System
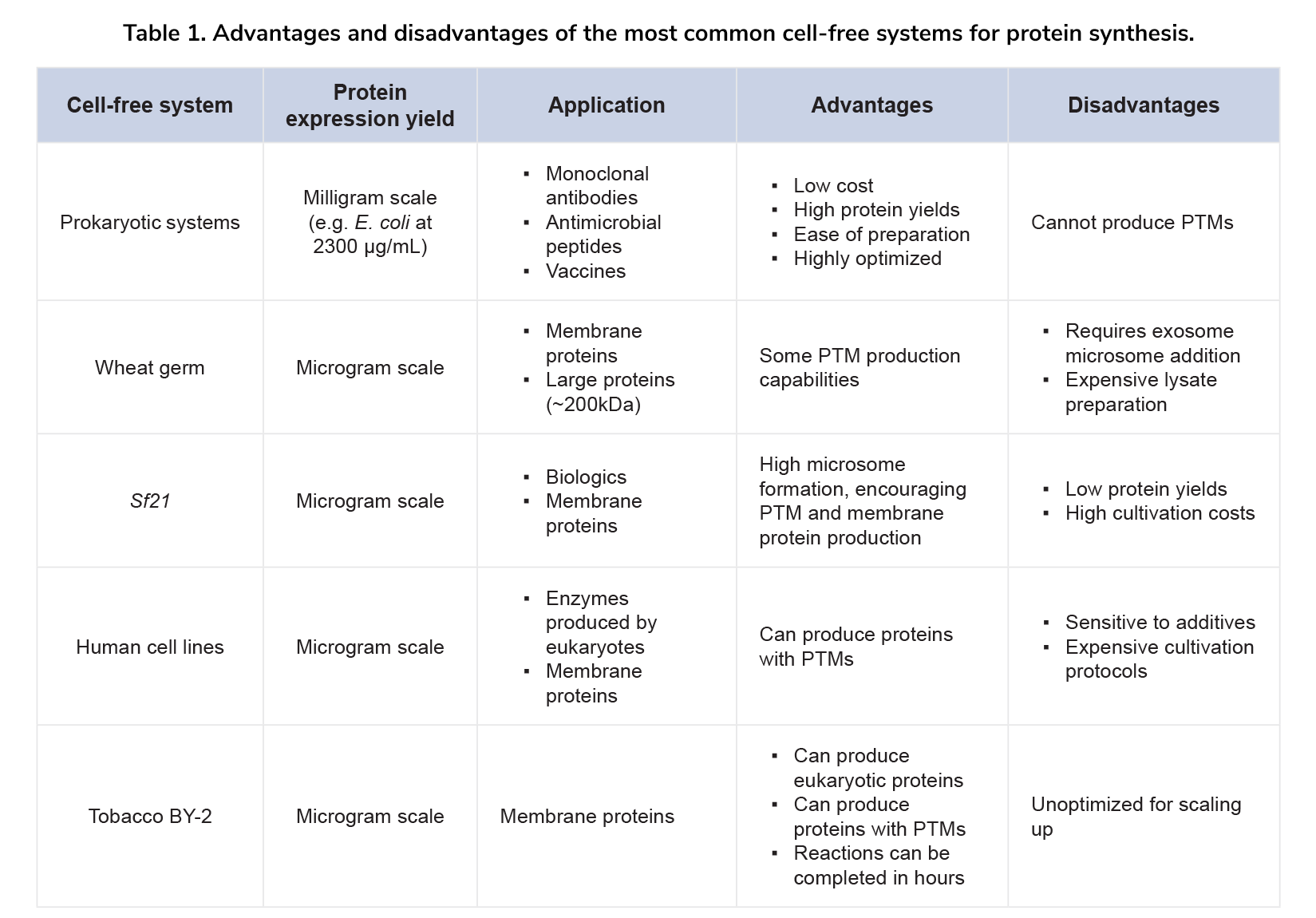
Like prokaryotic systems, eukaryotic CFPS systems have unique advantages and drawbacks that impact protein synthesis (Table 1). Each system's characteristics must be evaluated to select the best one for specific proteins. Here are key factors to consider when choosing a CFPS system:
- Protein Yield:
Different CFPS systems produce varying yields based on the lysates' biochemical composition. Systems that couple transcription and translation typically generate higher yields. Additionally, DNA structures, like a 5’-UTR with an enhancer sequence, can boost transcription and protein synthesis[24,25]. - Protein Type:
Different proteins require specific reagents and biomolecules. For example, integral membrane proteins need lipid bilayers or vesicles for purity and integrity[26]. Therapeutic proteins like onconases, which cleave tRNAs, require systems that pulse tRNA intake or maintain tRNA stability[27]. - Post-Translational Modifications:
Some CFPS systems, particularly prokaryotic-based and wheat-germ systems, cannot perform post-translational modifications. For producing monoclonal antibodies with disulfide bridges or glycosylated proteins, eukaryotic CFPS systems are more suitable[28].
Evolution of CFPS Systems
Currently available prokaryotic CFPS systems can produce a wide variety of biopharmaceutical proteins. However, each species has unique strains that can further enhance protein yields and quality. Prokaryotic systems, particularly E. coli, can be engineered to synthesize non-canonical amino acids using strains like C321.DA[29].
Eukaryotic CFPS systems can also be cloned to produce specific proteins at an industrial scale. Additionally, genomes from both prokaryotic and eukaryotic systems can be engineered to study biochemical pathways and assess protein interactions, driving R&D efforts[30,31].
CFPS systems offer diverse and versatile formats for synthesizing specific proteins, each with unique advantages and considerations. Understanding the landscape of prokaryotic and eukaryotic systems, as well as the key influencing factors such as protein type, yield, and post-translational modifications, is essential for optimizing protein synthesis.
The future of advanced CFPS systems looks bright, with genetic engineering addressing shortcomings of today’s systems by enhancing protein yields, quality, and functionality. Moreover, the integration of automation, artificial intelligence, and machine learning is further streamlining protein production, enabling precise control and high-throughput screening at unprecedented scales.
Work With Us
Accelerate your scientific discovery with the Tierra Protein Platform. Our platform integrates automation, computation, and high-throughput screening allowing you to synthesize custom proteins from diverse sources at the click of a button. Visit our ordering portal or contact Tierra Biosciences today to learn how our platform and expert guidance can drive the success of your programs.